Scientific Program

Ruchika
Japan Advanced Institute of Science and Technology, Japan
Title: Development of an artificial deaminase system with plant derived “DYW†type PPR protein
Biography:
Ruchika is perusing her doctoral degree in molecular biology under the supervision of Prof. Toshifumi Tsukahara at Japan Advanced institute of science and Technology (JAIST), Ishikawa, Japan. During her PhD, she is exploring her understanding on RNA editing in plants, bacterial cells as well as in mammalian cells. Ruchika did her master’s and bachelor’s in Botany (honors) from university of Delhi, India. Ruchika received the MEXT scholarship from Japanese government as well as other research grants for her outstanding research proposal for identifying the candidate genes related to U-to-C RNA editing in plants, especially hornworts. Very recently, she was honored by “Young scientist award†in Plant Genomics 2021
Abstract
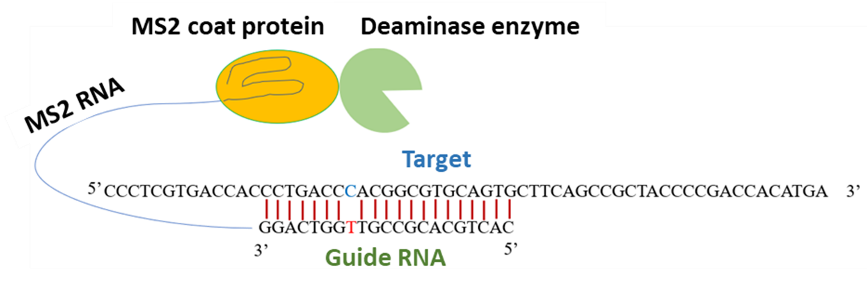
A genetic disorder is a disease caused by a change in the nucleotide base sequences. Many genetic diseases are raised due to T-to-C point mutations. Hence, editing of mutated genes represents a promising strategy for treating these disorders. In this study, an expression system was developed in which the PPR protein variants were cloned with PPR56 (truncated DYW), Physcomitrella patens (moss) editing factor. The assay system allowed to study RNA editing by PPR genes with its potential target sequences in mammalian cells. We engineered an artificial RNA editing mechanism by combining the deaminase domain of plant derived PPR56 (DYW) with a guideRNA (gRNA) which is complementary to target mRNA. In this artificial enzyme system, gRNA is bound to MS2 stem-loop, and deaminase domain, which can convert mutated target nucleotide C-to-U, is fused to MS2 coat protein. As a target RNA, we used RNA encoding blue fluorescent protein (BFP) which was derived from the gene encoding GFP by T>C mutation. Earlier MS2 system has been used with APOBEC1, showed 21% of GFP restoration. Upon transient expression of both components (DYW and gRNA), we confirmed the restoration of original sequence of mutated GFP revealing an editing efficiency of up to 85-100%. We successfully developed a bio-engineered RNA editing system using deaminase (PPR56DYW) in combination with MS2 system showing the maximum efficiency.
- Plant Genome Science
- Agribusiness
- Molecular Plant Breeding
- CRISPR-Cas9 system in plants
- Plant Genome Integrity
- Plant Biotechnology
- Plant Virology
- Medicinal Plants
- Plant Physiology
- Plant Genetics Market

